GUIskew
Installation
For linux and mac it can be installed via the pypi page IF you already have tkinter installed. On most Linux distros you can install tkinter by the following command:
$ apt-get install python-tk
Alternatively, for Windows, the exe can be downloaded here:
Usage
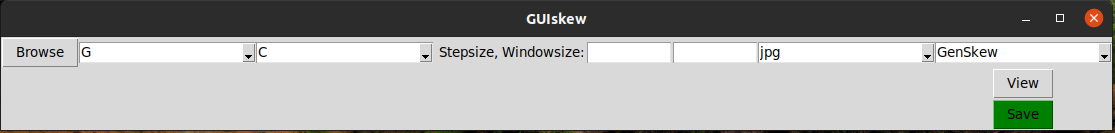
File Selection
When clicking the Browse button, a new window will appear in which you can select one or multiple files. To select gb or .gz files you will have to select the file type in the appropriate dropdown menu. After pressing open, the selected file or files should be displayed below.
Changing Settings
Next to the Browse button you can change the two nucleotides as well as the stepsize and windowsize and outputfiltype. The step and windowsize don't have to be specified and will be calculated automatically by dividing the sequence lenght by 1000 if left out.The step and windowsize are limited to 100. This is because if they are lower, there could be none of the specified nucleotides in the window and the formula used to calculate the skew would divide by 0. With the dropdown menu on the far right you can select between the normal GenSkew graph and the SkewIT¹ graph.
Viewing and Saving the Graphs
When pressing the View button the Graph will be displayed in the window, but NOT saved. This only works if only one sequence is selected. The Save button however saves the graph or graphs. Once pressed, a window will appear in which you can select the folder that everything is going to be saved in. If you press cancel, the graphs will be saved to wherever the sequencefiles are from.
References
1: SkewIT, https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1008439 (06.04.2022), SkewIT: The Skew Index Test for large scale GC Skew analysis of bacterial genomes, Jennifer Lu, Steven L. Salzberg
